paired end sequencing insert size
Velvet insert length on Illumina NGS Paired end reads. How to quickly check the insert size distribution of a new sequencing library.
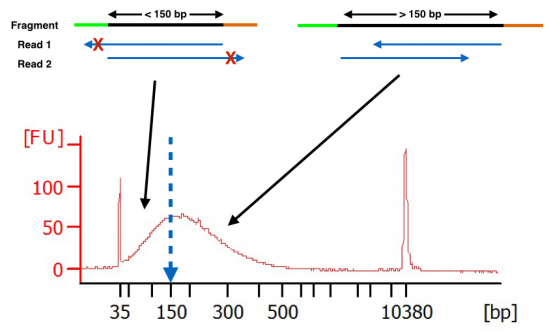
How Short Inserts Affect Sequencing Performance
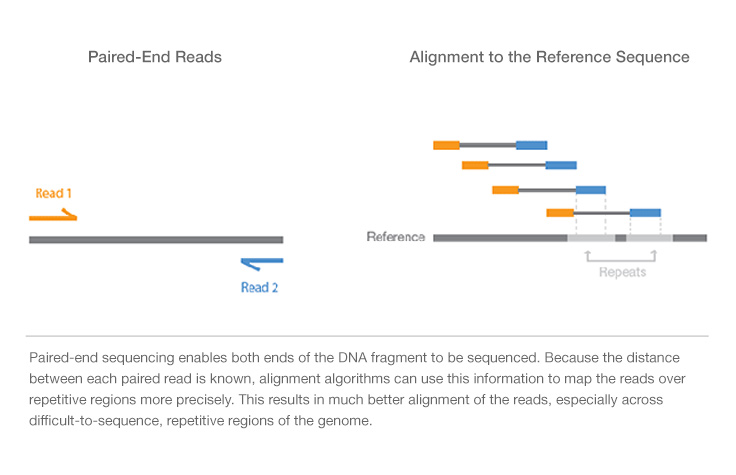
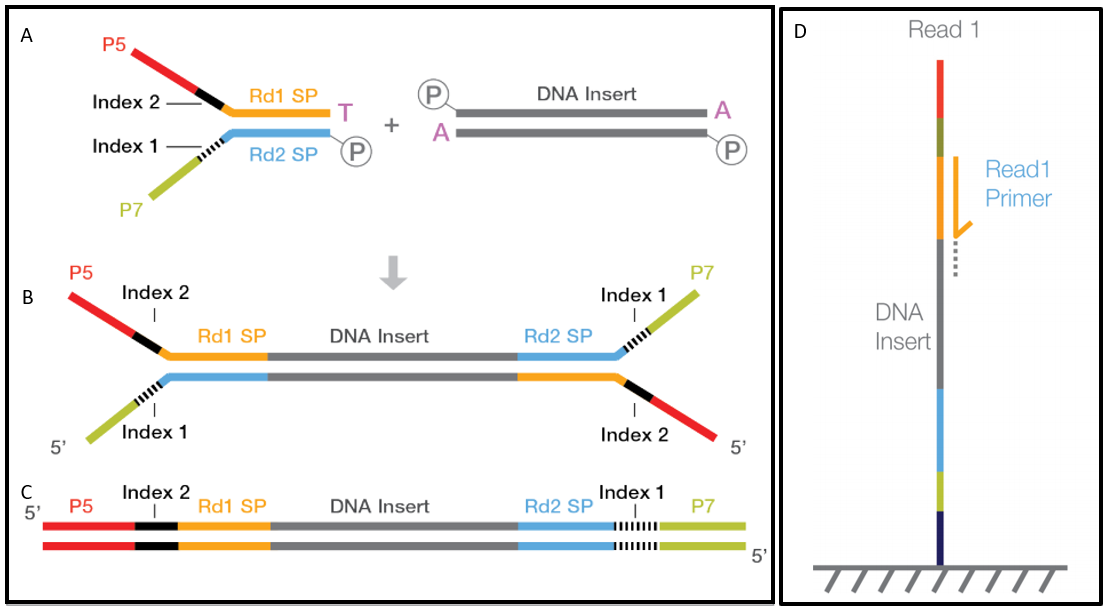
Basic notations of a paired end sequenced DNA fragment.
. Depending on the length of your reads it is possible for PE reads to have overlapping ends. Download scientific diagram Paired-end PE sequencing and insert size IS filtering to increase sensitivity. For Illumina systems DNA insert size range of 200800 bp.
Such reads will also contain adapter sequences which may need to be trimmed they as can negatively impact on some types of. A Sample-wise IS histograms of liquid biopsies LBs. Including flanking intronic regions of.
Thats why you should first take a look in the operating instructions of your used device and kit. By computational modeling we show how libraries of multiple insert sizes combined with strand-specific paired-end SS-PE sequencing can increase the information gained on alternative splicing especially in higher eukaryotes. However if it takes you 2 years to assemble and annotate the genome and only then you check this theres probably nothing that can be done.
Despite the benefits of gaining SS-PE data with paired ends of varying distance the standard Illumina protocol allows only non-strand-specific paired. The difference between BAC and fosmids is the size of the DNA inserted. In an Illumina sequencing run either single-end sequencing SE or paired-end sequencing PE can be used.
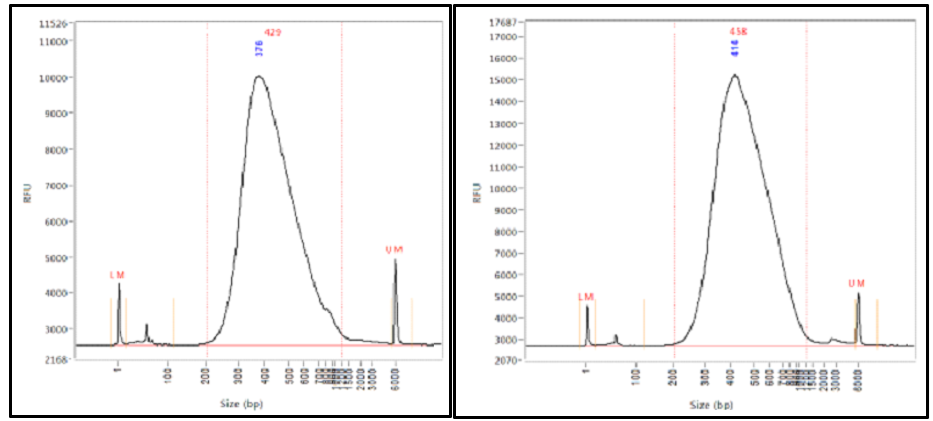
If your 10 kb mate pair library turns out to be mostly 300 bp inserts and you report this to your sequencing center a month after they give you your data chances are they can do something about it. Complete workflow automation system for NGS library prep for genomic sequencing. One could argue that longer read length generates more output from the same amount of input molecules therefore the insert size should be increased accordingly to make use of this extra data.
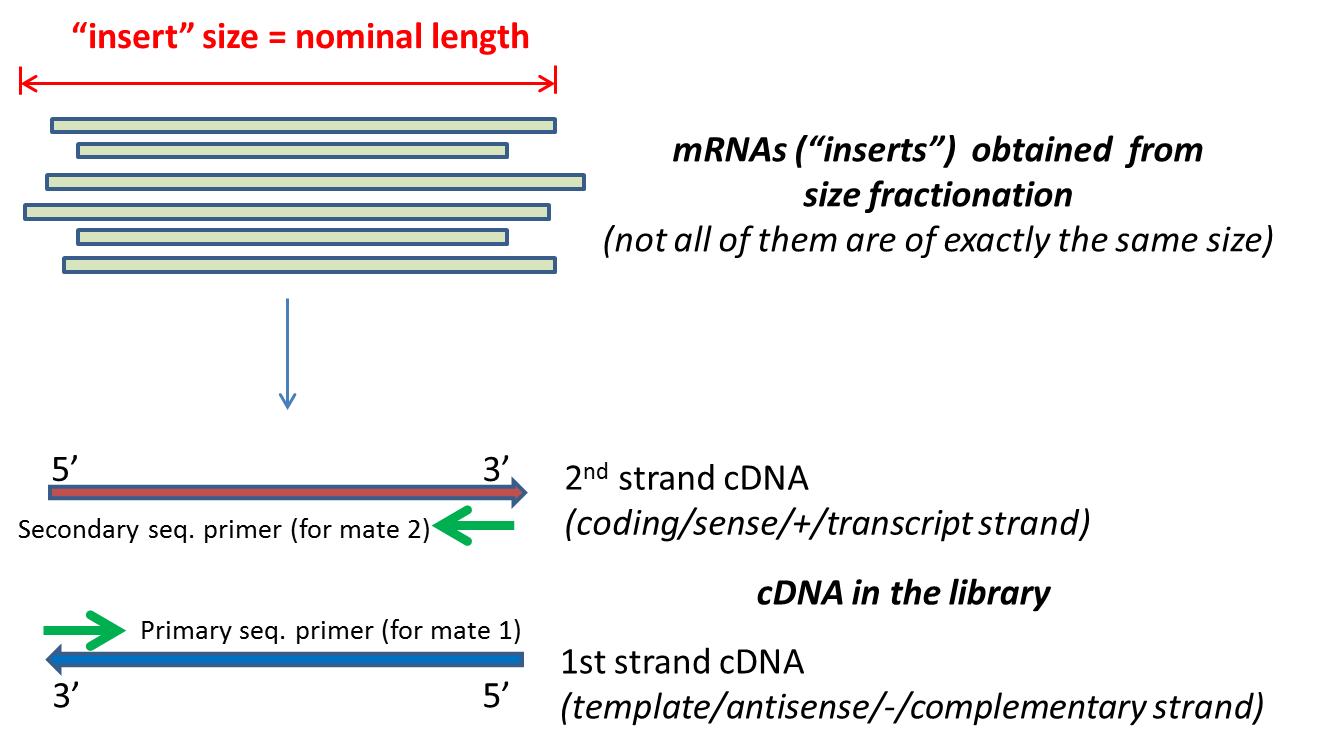
The size of fragments inserted is 150350 kb. You can use sequencing reagents to generate single continuous reads or for. Paired end sequencing refers to the fact that the fragment s sequenced were sequenced from both ends and not just the one as was true for first.
You can use sequencing reagents to generate single continuous reads or for paired-end sequencing in both directions. - either the enitre read dataset or a subset of it see below - a reference. For paired-end RNA-Seq use the following kits with an alternate fragmentation protocol followed by standard Illumina paired-end cluster generation and sequencing.
For instance for a PE150 sequencing run the insert size should be above 300bp. Ad Automate NGS library preparation lab automation technology from Automata. 2Align paired-end RNA-Seq data to the reference transcripts using minimum insert-length as zero and maximum insert-length as 500 or more.
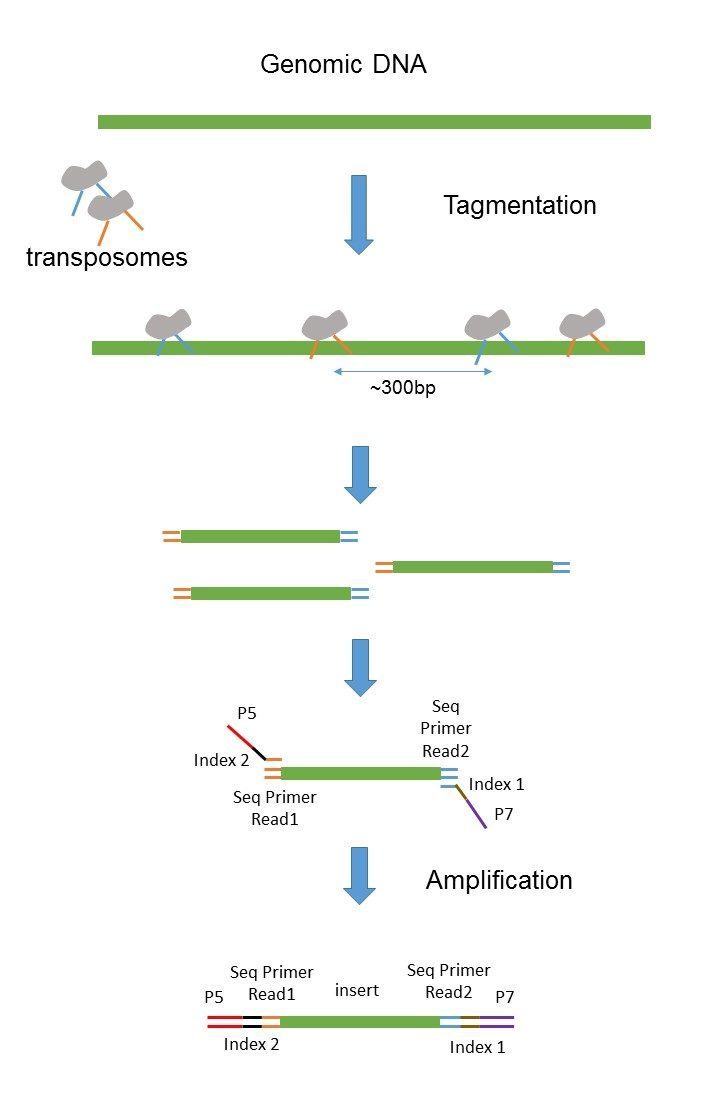
In any case DNA is chopped into small fragments ligated. For this reason there is no general recommendation and we can only give. For mRNA-Seq library prep use.
For paired-end RNA-Seq use the following kits with an alternate fragmentation protocol followed by standard Illumina paired-end cluster generation and sequencing. The difference in insert size stems from the difference in protocols. If they occur at significant levels the amount of useable sequence in a dataset will be reduced.
The insert is normally the stretch of sequence between the paired-end adapters so in your case the insert size would be 250 bp 2x75 bp reads 100 bp unsequenced middle piece. It depends on the used Illumina system the used reagents kits and the mode singlepaired end. B Histology-wise mean IS.
What is the optimal insert size. However the average size of human coding exons is only 160bp. Insert size for paired end sequencing for identification of structural variation.
In this blog post we will talk about what is the insert size in Illumina sequencing reads and how to find it in SAMBAM files and how to filter BAM files with it. For example a 300-cycle kit can be used for a 1 300 bp single-read run or a 2 150 bp paired-end run Beginners Guide to Next-Generation Sequencing. Thus the other ends can be used to fill the un-sequenced regions in the.
Smaller DNA fragments are aligned to the largest DNA fragments by one of their two-ends. There are probably several ways to do this but here is what we tend to use. Ranges from 2 150 to 2 300 bp.
Basically a complete NGS fragment prepared for sequencing consists of the original DNA fragment insert size and the adapters added at each end. Another commonly used artificial chromosome is fosmid. Despite the benefits of gaining SS-PE data with paired ends of varying distance the standard Illumina protocol allows only non-strand-specific paired-end sequencing with a single insert size.
1Create reference transcriptome index for BowTie or BWA. Regarding paired -end assembly insert-size calculation on newbler23. Paired-end RNA sequencing RNA-Seq enables discovery applications such as detecting gene fusions in cancer and characterizing novel splice isoforms.
Ad Automate NGS library preparation lab automation technology from Automata. Fosmids can only hold 40 kb DNA fragments which allows a more accurate breakpoint determination. Insert size -- The insert size refers to the distance between the pairs.
By paired-end sequencing of a series of stepwise insert size libraries we are able to recover the full length sequences of the largest DNA fragments using computational method. After you get the reads be they paired end or mate pairs from the sequencing centre you sometimes want to chek thr distribution of the insert sizes. Firstly they allow an orientation-specific binding of the complete fragment to the flow cell by hybridization.
PE reads generally have a smaller insert size 1kp than MP 2-5 kb. The adapters have two main functions. However the average size of.
-I 0 -X 500 BWA. Complete workflow automation system for NGS library prep for genomic sequencing. Paired end reads with an insert size of less than the length of a single read contain less information than read pairs with longer insert sizes.
PE reads generally have a smaller insert size 1kp than MP 2-5 kb.

What Is Mate Pair Sequencing For

Interpreting Color By Insert Size Integrative Genomics Viewer

Diagram To Show The Construction Of A Fragment With An Insert Size Is Download Scientific Diagram

Definition Of Essential Terms A Definition Of Fragment Read And Download Scientific Diagram

What Are Paired End Reads The Sequencing Center

Pdf Assessment Of Insert Sizes And Adapter Content In Fastq Data From Nexteraxt Libraries

Library Insert Size Libraries Were Constructed Using The Standard A Download Scientific Diagram
What Read Lengths Are Produced By Modern Illumina Sequencers
The Insert Size In Paired End Data Seqanswers
What Is Paired End 150 Omega Bioservices

Interpreting Color By Insert Size Integrative Genomics Viewer

What Are Paired End Reads The Sequencing Center

What Is Mate Pair Sequencing For
How Short Inserts Affect Sequencing Performance
What Happens When Hardware Advances Faster Than Molecular Protocols Cofactor Genomics

Diagram To Show The Construction Of A Fragment With An Insert Size Is Download Scientific Diagram